A Simple Note for Gaussian Mixture Models
1. Gaussian Mixture Model
A Gaussian mixture is a probabilistic model that assumes all the data points are generated from a mixture of a finite number of Gaussian distributions with unknown parameters.
Gaussian mixture model(GMM) is an clustering algorithm similar to k-means. Unlike k-means, GMM is a soft clustering, each instance associates with a cluster with a probability. Formally, given a set of observations \(X = (x_1, x_2, ..., x_n)\), \(x_i \in R^{d}\), and \(i=1, 2, ..., n\), Gaussian mixture model assumes:
-
there are \(K\) mixture components, component value denoted as \(k \in \{1, 2, ..., K\}\)
-
\(x_i\) comes from component \(z_i\), \(Z = (z_1, z_2, ..., z_n)\) is the latent variables
-
\(\begin{equation} P(z_i=k) = \pi_k \end{equation}\), s.t. \(\sum_{k=1}^K \pi_k = 1\), \(\pi = (\pi_1, \pi_2, ...\pi_K)\) is the mixing proportions
-
\(\begin{equation} P(x_i=x | z_i=k) \sim N(\mu_k, \Sigma_k) \end{equation}\), \(N(\mu_k, \Sigma_k)\) is a multivariate Gaussian distribution, the density function is
where \(\Sigma_k\) is the \(d \times d\) covariance matrix for component \(k\). The objective of GMM is to estimate the probabilities that each instance belongs to each component, that is
\[\begin{equation} P(z_i=k | x_i), k = 1, 2, ... K, i=1, 2, ... n \label{obj} \end{equation}\]2. Bayes and Maximum Likelihood Estimation
With Bayes rule, we have
\[\begin{equation} \begin{aligned} P(z_i=k|x_i) & = \frac{P(x_i|z_i)P(z_i=k)}{P(x_i)} \\ & = \frac{P(x_i|z_i)P(z_i=k)}{\sum_{k=1}^KP(x_i|z_i)P(z_i=k)} \\ & = \frac{\pi_k*N(x_i, \mu_k, \Sigma_k)}{\sum_{k=1}^K\pi_k*N(x_i, \mu_k, \Sigma_k)} \\ & = w_{ik} \end{aligned} \label{weights} \end{equation}\]If we know parameters \(\theta = (\pi_1, \pi_2, ..., \pi_K, \mu_1, \mu_2, ..., \mu_K, \Sigma_1, \Sigma_2, ..., \Sigma_K)\), we will get the objective. Therefore, the target becomes to estimate the unknown parameter \(\theta\). To estimate \(\theta\), GMM uses maximum likelihood estimation(MLE),
\[\begin{equation} \begin{aligned} L(\theta|X, Z) & = P(X, Z|\theta) \\ & = \prod_{i=1}^n \sum_{k=1}^K P(x_i=x|z_i=k)P(z_i=k) \\ & = \prod_{i=1}^n \sum_{k=1}^K \pi_k N(\mu_k, \Sigma_k) \end{aligned} \end{equation}\]log-likelihood becomes
\[\begin{equation} \begin{aligned} l(\theta|X, Z) & = \sum_{i=1}^n log(\,\sum_{k=1}^K \pi_k N(\mu_k, \Sigma_k))\, \end{aligned} \end{equation}\]Normally, we can get
\[\begin{equation} \theta^{*} = \underset{\theta}{\operatorname{argmax}} l(\theta|X, Z) \end{equation}\]by set
\[\begin{equation} \frac{\partial l}{\partial \theta} = 0 \end{equation}\]However, it’s hard to solve \(\theta\) with closed-form expressions now. So GMM uses the Expectation—maximization algorithm(EM).
3. Expectation—Maximization Algorithm
In statistics, an expectation–maximization (EM) algorithm is an iterative method to find (local) maximum likelihood or maximum a posteriori (MAP) estimates of parameters in statistical models, where the model depends on unobserved latent variables.
EM iterates the expectation step(E-step) and the maximization step(M-step) until converge:
- initializes \(\theta = (\pi_1, \pi_2, ..., \pi_K, \mu_1, \mu_2, ..., \mu_K, \Sigma_1, \Sigma_2, ..., \Sigma_K)\) to \(\theta_0\)
-
start iteration until convergence or reach the max iterate number, at step \(t\geq0\):
2.1 E-step: with \(\theta_{t}\), calculates \(\begin{equation} P(z_i=k|x_i) \end{equation}\) through equation \(\eqref{weights}\) for \(k=1, 2, ..., K\) and \(i=1, 2, ..., n\)
2.2 M-step: estimates \(\theta_{t+1}\) by Maximum Likelihood Estimation
In E-step, EM defines the expected value of the log-likelihood function of \(\theta\) as
\[\begin{equation} \mathbf{Q}(\theta|\theta_{t}) = \mathbf{E}_{Z|X, \theta^t}(logL(\theta;X,Z)) \end{equation}\]In M-step, EM estimates next \(\theta\) as
\[\begin{equation} \theta^{t+1} = \underset{\theta}{\operatorname{argmax}} \mathbf{Q}(\theta|\theta_{t}) \quad s.t. \ \sum_{k=1}^K \pi_k = 1 \end{equation}\]4. GMM with EM
In GMM situation, we have
\[\begin{equation} \begin{aligned} L(\theta;x_i, z_i) & = \prod_{k=1}^K I(z_i=k) P(x_i, z_i|\theta) \\ & = \prod_{k=1}^K I(z_i=k) P(x_i|z_i, \theta) P(z_i | \theta) \\ & = \prod_{k=1}^K I(z_i=k) N(x_i|\mu_k, \Sigma_k) \pi_k \end{aligned} \end{equation}\]where \(I(z_i=k)\) is a indicator function. And
\[\begin{equation} L(\theta|X, Z) = \prod_{i=1}^n L(\theta;x_i, z_i) \end{equation}\]In E-step, the expected value of the log-likelihood function becomes
\[\begin{equation} \begin{aligned} \mathbf{Q}(\theta|\theta_{t}) & = \mathbf{E}_{Z|X, \theta^t}(logL(\theta;X,Z)) \\ & = \mathbf{E}_{Z|X, \theta^t}\sum_{i=1}^nlog(L(\theta;x_i, z_i)) \\ & = \sum_{i=1}^n \mathbf{E}_{z_i|x_i, \theta^t} log(L(\theta;x_i, z_i)) \\ & = \sum_{i=1}^n \mathbf{E}_{z_i|x_i, \theta^t} \sum_{k=1}^K I(z_i=k) *(log\pi_k + logN(x_i|\mu_k, \Sigma_k)) \\ & = \sum_{i=1}^n \sum_{k=1}^K P(z_i=k|x_i) * (log\pi_k + logN(x_i|\mu_k, \Sigma_k)) \end{aligned} \end{equation}\]Combine the current \(\theta_{t}\) and equation \(\eqref{weights}\), we have
\[\begin{equation} w_{ik} = P(z_i=k|x_i) \end{equation}\]where \(w_{ik}\) is a constant. Thus
\[\begin{equation} \mathbf{Q}(\theta|\theta_{t}) = \sum_{i=1}^n \sum_{k=1}^K w_{ik} * (log\pi_k + logN(x_i|\mu_k, \Sigma_k)) \end{equation}\]In M-step, the optimal problem becomes
\[\begin{equation} \begin{aligned} \underset{\theta}{\operatorname{max}} \quad & \mathbf{Q}(\theta|\theta_{t}) \\ \text{s.t.} \quad & \sum_{k=1}^K \pi_k = 1 \end{aligned} \end{equation}\]which is a Lagrangian problem. Therefore,
\[\begin{equation} \begin{aligned} H(\theta, \lambda) = \mathbf{Q}(\theta|\theta_{t}) + \lambda(\sum_{k=1}^K \pi_k - 1) \end{aligned} \end{equation}\]Take partial derivatives, we have
\[\begin{equation} \begin{aligned} \frac{\partial H}{\partial \mu_k} & = \sum_{i=1}^n w_{ik} * \frac{\partial logN(x_i|\mu_k, \Sigma_k)}{\partial \mu_k} \\ & = \sum_{i=1}^n w_{ik} \Sigma_{k}^{-1}(x_i - \mu_k) \end{aligned} \label{partialmu} \end{equation}\]and
\[\begin{equation} \frac{\partial H}{\partial \Sigma_k} = \frac{1}{2}\{ \sum_{i=1}^n w_{ik} \Sigma_{k}^{-T} (x_i - \mu_k)(x_i-\mu_k)^{T}(\Sigma^{-T} - I) \} \label{partialSigma} \end{equation}\]and
\[\begin{equation} \frac{\partial H}{\partial \pi_k} = \sum_{i=1}^n w_{ik} / \pi_k + \lambda \label{pi_k} \end{equation}\]and
\[\begin{equation} \frac{\partial H}{\partial \lambda} = \sum_{k=1}^K \pi_k - 1 \label{lambda} \end{equation}\]Set \(\eqref{partialmu}\) equal to zero, we have
\[\begin{equation} \mu_k^* = \frac{\sum_{i=1}^n w_{ik}x_i}{\sum_{i=1}^n w_{ik}} \end{equation}\]Set \(\eqref{partialSigma}\) equal to zero, and combine with \(\mu_k^*\), we have
\[\begin{equation} \Sigma_k^* = \frac{\sum_{i=1}^n w_{ik}(x_i-\mu_k)(x_i-\mu_k)^{T} }{\sum_{i=1}^n w_{ik}} \end{equation}\]Set \(\eqref{pi_k}\) and \(\eqref{lambda}\) equal to zero, we have
\[\begin{equation} \pi_k^* = \frac{\sum_{i=1}^n w_{ik}}{\sum_{k=1}^K \sum_{i=1}^n w_{ik}} \end{equation}\]Therefore, we got \(\theta_{t+1} = (\mu_1^*, \mu_2^*, ... \mu_K^*, \Sigma_1^*, \Sigma_2^*, ... \Sigma_K^*, \pi_1^*, \pi_2^*, ... \pi_K^*)\).
5. A numpy python implementation
First, we need to initialize \(\theta_0\). Here, we use kmeans++, kmeans and random to initialize parameters. After initialization, we calculate the weights equation \(\eqref{weights}\), which is the E-step.
# 0. import
import numpy as np
from sklearn.datasets import make_blobs
from sklearn.cluster import KMeans
import matplotlib.pyplot as plt
from scipy.stats import multivariate_normal
X, y = make_blobs()
# 1. EM: Initlization and E-step
def _kmeans_init(X, n_component, kmeans_method):
n_samples = X.shape[0]
if kmeans_method == 'kmeans++':
kmeans = KMeans(n_clusters=n_components).fit(X)
else:
kmeans = KMeans(n_clusters=n_components, init='random').fit(X)
labels = kmeans.labels_
weight_matrix = None
for i in range(n_component):
sub_component = X[labels==i]
pi = sub_component.shape[0]/n_samples
mean = np.mean(sub_component, axis=0)
cov = np.cov(sub_component.T)
pdf_value = multivariate_normal.pdf(X, mean, cov)
weight = pi*pdf_value
if weight_matrix is None:
weight_matrix = weight
else:
weight_matrix = np.column_stack((weight_matrix, weight))
weight_sum = np.sum(weight_matrix, axis=1)[:, np.newaxis]
weights = weight_matrix/weight_sum
return weights
def _initialize(X, n_component, method):
# default random
n_samples = X.shape[0]
if method == 'kmeans++':
weights = _kmeans_init(X, n_component, 'kmeans++')
elif method == 'kmeans_random':
weights = _kmeans_init(X, n_component, 'kmeans_random')
else:
weights = np.random.rand(n_samples, n_components)
w = weights.sum(axis=1)
weight_sum = weights.sum(axis=1)[:, np.newaxis]
weights /= weight_sum
return weights
Given the value of weights equation \(\eqref{weights}\), we update the value of \(\theta\) through MLE, which is the M-step. To avoid forloop, convert the numerator of equation \(\eqref{partialSigma}\) as:
\[\begin{equation} \begin{aligned} A & = \sum_{i=1}^n w_{ik}(x_i - \mu_k)(x_i - \mu_k)^T \\ \Rightarrow \quad A_{st} &= \sum_{i=1}^n w_{ik}x_{is}x_{it} - \sum_{i=1}^nw_{ik}\mu_{ks}x_{it} - \sum_{i=1}^nw_{ik}\mu_{kt}x_{is} + \sum_{i=1}^nw_{ik}\mu_{ks}\mu_{kt} \\ \Rightarrow \quad A_{st} & = A1_{st} - A2_{st} - A3_{st} + A4_{st} \end{aligned} \end{equation}\]for \(1 \leq s, t \leq d\), where
\[\begin{equation} \begin{aligned} A1 & = w_k*(X.T) \\ A2 & = A3.T \\ A3 & = \mu_k((w_k*(X.T)).dot(H)) \\ A4 & = (\sum_{i=1}^n w_{ik})*(\mu_k.dot(\mu_k.T)) \\ w_k & = np.array([w_k1, w_k2, ... w_kn]) \\ \mu_k & = np.array([\mu_k1, \mu_k2, ..., \mu_kd]) \\ H & = np.ones(shape=(n, d)) \end{aligned} \end{equation}\]# 2. EM: M-step
def _update_means(X, weights):
weight_sum = np.sum(weights, axis=0)
means = X.T.dot(weights)/weight_sum
return means
def _update_covs(X, n_components, weights, means):
n, m = X.shape
covs = None
H = np.ones(shape=(n, m))
for k in range(n_components):
weight = weights[:, k]
weight_sum = np.sum(weight)
mean = means[:, k]
weighted_X_T = weight*(X.T)
a1 = (weighted_X_T).dot(X)
a3 = mean*(weighted_X_T.dot(H))
a2 = a3.T
mean_reshaped = mean.reshape(-1, 1)
a4 = weight_sum*((mean_reshaped).dot(mean_reshaped.T))
a = a1 - a2 - a3 + a4
cov = a/weight_sum
if covs is None:
covs = cov
else:
covs = np.dstack((covs, cov))
return covs
def _update_pis(weights):
w_each_component = np.sum(weights, axis=0)
w_sum = np.sum(w_each_component)
pis = w_each_component/w_sum
return pis
def _update_weights(X, n_components, means, covs, pis):
weighted_pdf_array = None
for k in range(n_components):
mean = means[:, k]
cov = covs[:,:,k]
pi = pis[k]
pdf_values = multivariate_normal.pdf(X, mean, cov)
weighted_pdf_values = pi*pdf_values
if weighted_pdf_array is None:
weighted_pdf_array = weighted_pdf_values
else:
weighted_pdf_array = np.column_stack((weighted_pdf_array, weighted_pdf_values))
weighted_sum = np.sum(weighted_pdf_array, axis=1)[:, np.newaxis]
weights = weighted_pdf_array/weighted_sum
return weights
# 3. gmm fit
def gmm_fit(X, n_components, init_method, max_iter=100):
weights = _initialize(X, n_components, init_method)
iter_left = max_iter
while iter_left > 0:
means = _update_means(X, weights)
covs = _update_covs(X, n_components, weights, means)
pis = _update_pis(weights)
weights_next = _update_weights(X, n_components, means, covs, pis)
if np.allclose(weights, weights_next):
print('converage at step', max_iter-iter_left)
break
else:
weights = weights_next
iter_left -= 1
return means, covs, weights_next
# 4. predict labels
def gmm_predict(X, n_components, means, covs, weights):
pdf_values = None
for k in range(n_components):
mean = means[:, k]
cov = covs[:, :, k]
pdf_value = multivariate_normal.pdf(X, mean, cov)
if pdf_values is None:
pdf_values = pdf_value
else:
pdf_values = np.column_stack((pdf_values, pdf_value))
labels = np.argmax(pdf_values, axis=1)
return labels
# 5. gmm
def gmm_fit_predict(X, n_components, init_method, max_iter):
means, covs, weights = gmm_fit(X, n_components, init_method, max_iter)
labels = gmm_predict(X, n_components, means, covs, weights)
return labels
# 6. result and demonstration
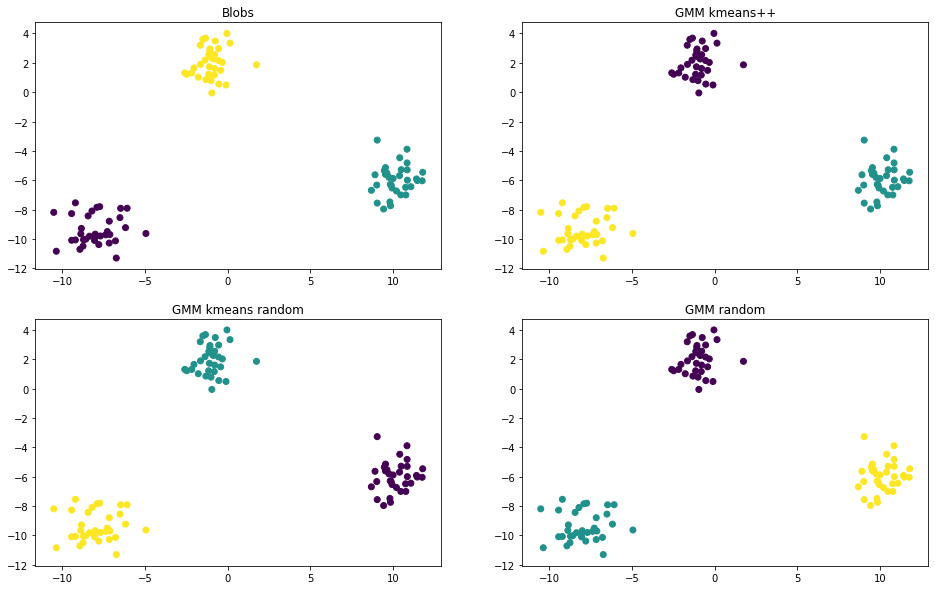
def plot_scatter(ax, X, labels, title):
ax.scatter(X[:, 0], X[:, 1], c=labels)
ax.set_title(title)
n_components = 3
gmm_labels_kmeans_plus_plus = gmm_fit_predict(X, n_components, 'kmeans++', max_iter=1000)
gmm_labels_kmeans_random = gmm_fit_predict(X, n_components, 'kmeans_random', max_iter=1000)
gmm_labels_random = gmm_fit_predict(X, n_components, 'random', max_iter=1000)
fig, axes = plt.subplots(2, 2, figsize=(16, 10))
plot_scatter(axes[0, 0], X, y, 'Blobs')
plot_scatter(axes[0, 1], X, gmm_labels_kmeans_plus_plus, 'kmeans++')
plot_scatter(axes[1, 0], X, gmm_labels_kmeans_random, 'kmeans random')
plot_scatter(axes[1, 1], X, gmm_labels_random, 'random')